 CS5131 Introduction to Artificial Intelligence
CS5131 Introduction to Artificial Intelligence
 CS5131 Introduction to Artificial Intelligence
CS5131 Introduction to Artificial Intelligence
BY LIEW WEI PYN AND PRANNAYA GUPTA
M22504
Done as part of CS5131: Introduction to Artificial Intelligence
The UV Aerosol Index (UVAI), which is also often referred to as the Absorbing Aerosol Index (AAI) or the residue, the spectral contrast anomaly, or simply aerosol index, is a reading measured based on wavelength-dependent changes in molecular Rayleigh Scattering, surface reflection, gaseous absorption and aerosol and cloud scattering in the UV spectral range for a pair of wavelengths, well known as the spectral contrast at the two wavelengths. The difference between observed and modelled reflectance results in the UVAI. When the UVAI is positive, it indicates the presence of elevated aerosols that absorb ultraviolet (UV) radiation in the atmosphere, such as dust and smoke.
These absorbing aerosols are solid and liquid particles suspended in the atmosphere. They are made up of primary aerosols and secondary aerosols. Primary aerosols are emitted directly, whereas secondary aerosols are formed by gases reacting in the atmosphere. The main sources of aerosols are combustion, like burning biomass, and natural sources, like volcanoes. The UVAI can show dust and smoke particles coming from events like dust storms and outbreaks, volcanic eruptions, fires, and biomass burning around the world, and hence it is useful for tracking the evolution of episodic aerosol plumes from such events. The wavelengths used have very low ozone absorption, so unlike aerosol optical thickness measurements, UVAI can be calculated in the presence of clouds. Daily global coverage is therefore possible.
Traditionally, aerosol optical thickness measurements are made using spaceborne sensors operating in the visible and infrared ranges of electromagnetic radiation, where multiple scattering in the atmosphere is less important than in the ultraviolet radiation and inversion calculations are relatively easier. In the visible and near-infrared waves, the large surface albedos of many land types make retrieval of aerosols difficult over these regions. With the ongoing development of numerical radiative transfer codes and increasing computational speeds, accounting for multiple scattering is no longer a problem, allowing for new techniques of aerosol measurements in the UV. Because the surface albedos of both land and ocean are small in the UV, this wavelength range should be suitable for aerosol detection over land.
A common way of measuring the UVAI is the Residue Method, where we simplify some wavelength-dependent variable, $r$ to denote UVAI. From here, we have the following expression:
$$r_\lambda = -100 \left[\log \left(\frac{I(\lambda)}{I\left(\lambda_0\right)}\right)_{measured} - \log \left(\frac{I(\lambda)}{I\left(\lambda_0\right)}\right)_{ray} \right]$$where $I(\lambda)$ is the radiance at the top of the atmosphere (TOA) at a wavelength $\lambda$. The subscript $measured$ refers to a measured TOA radiance of a real atmosphere with aerosols, as opposed to a calculated TOA radiance for an aerosol-free atmosphere with only Rayleigh scattering and absorption by molecules and surface reflection and absorption, which is referred to as $ray$ in above equation. $\lambda_0$, on the other hand, is the wavelength of reference when computing the UV Aerosol Index at different wavelengths. For instance, in the case of the Nimbus 7 Total Ozone Mapping Spectrometer (TOMS) for the years 1979-1993, we noted the use of the 340 and the 380 nm radiances as $I(\lambda)$ and $I(\lambda_0)$ respectively, and hence the computation or $r_\lambda$, thereby referred to as $\Delta N_\lambda$ in Herman et al, is as follows:
$\Delta N_\lambda = -100 \left(\log \left(\frac{I_{340}}{I_{380}} \right)_{meas} - \log \left(\frac{I_{340}}{I_{380}} \right)_{ray} \right)$
This is a general form of the computation. However, this is not sufficient extrapolation of this task. We can simplify this further using the concepts of reflectance and the surface albedo.
We first note reflectance, defined as follows:
$R = \frac{\pi I}{E_0 \cos \left(\theta_0\right)}$
Here, $I$ is similarly a function of $\lambda$, similarly referring to radiance at the TOA, while $E_0$ depicts the solar irradiance at TOA perpendicular to the direction of the incident sunlight. $\theta_0$ depicts the Solar Zenith Angle, as shown below:
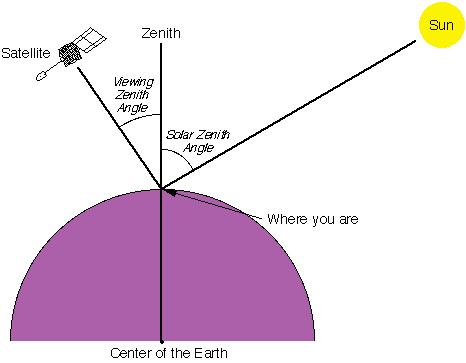
Source: Support to Aviation Control Service (SACS) Article on the Solar Zenith Angle
We note that the TOA plane at which $E_0$ is measured is perpendicular to the beam ray which comes at $\theta_0$. Hence, it is necessary to adjust $E_0$ to be altered by this beam, giving the $E_0$ about the TOA plane perpendicular to the ground surface, which is to be measured. This gives the solar irradiance about the observer's TOA, which is divides by his solar radiance, allowing us to get an accurate value for the reflectance. From here, we have another representation of $I(\lambda)$, which continues to simplify the above result by introducing this term know as the surface albedo.
The surface albedo $A_s$ for the Rayleigh atmosphere calculation is chosen so that
$R_{\lambda_0, meas} = R_{\lambda_0, ray} (A_s)$
$r_\lambda = -100 \left(\log R_{\lambda, meas} - \log R_{(A_s)\lambda_0, ray} \right)$
This is a useful mathematical formula, since the simulation is not problematic to compute for near-range satellites such as the Sentinel-5.
In this project, we aim to train a model to predict the UVAI readings, based on the ambient gaseous concentrations of specific gases in the atmosphere, including Methane (CH4), Sulphur Dioxide (SO2), Ozone (O3), Nitrogen Dioxide (NO2), Formaldehyde (HCHO) and Carbon Monoxide (CO).
We specifically train this on data from the United States of America (USA), specifically segmented for the specific states in the country. This is also categorical data which we will convert to discrete. This data is acquired from the Google Earth Engine based on the data from the European Space Agency's (ESA) Sentinel-5 Precursor (S5p) Mission Satellite, a Low-Earth Orbit Polar Satellite System part of the Global Monitoring of the Environment and Security (GMES/COPERNICUS) space component programme headed by the European Commission (EC) in partnership with the aforementioned ESA. The goal of the Satellite is to provide information and services on air quality, climate, and the ozone layer. The S5p mission includes the TROPOspheric Monitoring Instrument (TROPOMI), which takes daily (mostly Near Real-Time) global observations of key atmospheric components, such as absorbing aerosols and gaseous concentrations in the environment, at a 5.5 x 3.5 kilometer (km) resolution.
The Sentinel-5 Precursor Satellite only contains readings for half of the Earth at any one time, hence we wish to use the models to predict the UV Aerosol Index readings on the other hemisphere of the Earth, since this can help us form a reasoning as to how the UV Aerosol Index is changing without gaps in the data.
Please note that a lot of this data is derived from the Google Earth Engine Dataset, and a lot of the code requires you to use the Google Earth Engine Library on Google Colab. We have also saved the retrieved data into this Google Drive link in Public Access (please do not share). Save these files locally in a directory data/.
!pip install geopandas
!pip install vaex
!pip install --upgrade ipython
!pip install blake3==0.2.1
from IPython.display import clear_output
clear_output()
Important note: remember to restart kernel after pip installing vaex, otherwise there will be error when importing vaex
from glob import glob
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn, json, vaex
from IPython.display import clear_output, HTML
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression, Ridge, Lasso
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error, mean_absolute_error, mean_squared_log_error, median_absolute_error, mean_absolute_percentage_error
from os import path
import ee
import geopandas
We now authenticate earth engine to retrieve data from the website. Note that this requires a google earth engine authenticated account. This is not needed if not rerunning the section with code cells for data collection as the source data will also be included, however the code for data retrieval is still included for rigorous verification purposes.
# Trigger the authentication flow.
ee.Authenticate()
# Initialize the library.
ee.Initialize()
DataHub's Natural Earth Polygons GeoJSON Dataset is a geodata data package geojson polygons for the largest administrative subdivisions in every country. The data comes from Natural Earth, a community effort to make visually pleasing, well-crafted maps with cartography or GIS software at small scale.
We have opened this dataset as a geopandas.GeoDataFrame object in countries_geojson, as shown below.
countries_geojson = geopandas.read_file("https://datahub.io/core/geo-ne-admin1/r/admin1.geojson")
countries_geojson
| ISO3166-1-Alpha-3 | country | id | name | geometry | |
|---|---|---|---|---|---|
| 0 | ABW | Aruba | 5150 | Aruba | POLYGON ((-69.99694 12.57758, -69.93639 12.531... |
| 1 | AFG | Afghanistan | 1741 | Badghis | POLYGON ((64.30624 35.39722, 64.32468 35.40177... |
| 2 | AFG | Afghanistan | 1742 | Hirat | POLYGON ((61.36393 35.59824, 61.36548 35.59850... |
| 3 | AFG | Afghanistan | 1743 | Bamyan | POLYGON ((67.74391 35.44342, 67.75476 35.44412... |
| 4 | AFG | Afghanistan | 1744 | Balkh | POLYGON ((67.25913 37.18515, 67.28145 37.18866... |
| ... | ... | ... | ... | ... | ... |
| 4642 | ZWE | Zimbabwe | 529 | Manicaland | POLYGON ((33.01165 -17.38399, 32.99745 -17.404... |
| 4643 | ZWE | Zimbabwe | 530 | Matabeleland South | POLYGON ((29.43994 -19.87930, 29.45699 -19.874... |
| 4644 | ZWE | Zimbabwe | 531 | Bulawayo | POLYGON ((28.49757 -20.06270, 28.50532 -20.062... |
| 4645 | ZWE | Zimbabwe | 532 | Masvingo | POLYGON ((31.04181 -19.25226, 31.19870 -19.248... |
| 4646 | ZWE | Zimbabwe | 533 | Mashonaland West | POLYGON ((30.01065 -15.64623, 30.05024 -15.640... |
4647 rows × 5 columns
Now, we consider datasets that specifically contain the United States of America, giving all 51 states, includeing Massachusetts and California, but excluding Alaska, Rhode Island, District of Columbia and Hawaii, since they stray from mainland USA. We save this in the us_geojson object.
us_geojson = countries_geojson[countries_geojson.iloc[:, 0] == "USA"]
us_geojson
| ISO3166-1-Alpha-3 | country | id | name | geometry | |
|---|---|---|---|---|---|
| 4407 | USA | United States of America | 3513 | Massachusetts | MULTIPOLYGON (((-70.80176 41.24808, -70.81090 ... |
| 4408 | USA | United States of America | 3514 | Minnesota | POLYGON ((-95.10282 49.35394, -94.98252 49.356... |
| 4409 | USA | United States of America | 3515 | Montana | POLYGON ((-104.04757 48.99262, -104.04623 48.8... |
| 4410 | USA | United States of America | 3516 | North Dakota | POLYGON ((-97.22609 48.99267, -97.22922 48.968... |
| 4411 | USA | United States of America | 3517 | Hawaii | MULTIPOLYGON (((-155.60652 20.13796, -155.5863... |
| 4412 | USA | United States of America | 3518 | Idaho | POLYGON ((-114.75136 46.71429, -114.74362 46.7... |
| 4413 | USA | United States of America | 3519 | Washington | MULTIPOLYGON (((-123.31566 46.14854, -123.3379... |
| 4414 | USA | United States of America | 3520 | Arizona | POLYGON ((-109.04662 36.99981, -109.04665 36.8... |
| 4415 | USA | United States of America | 3521 | California | MULTIPOLYGON (((-118.59569 33.03528, -118.5898... |
| 4416 | USA | United States of America | 3522 | Colorado | POLYGON ((-103.66447 40.99974, -103.54540 40.9... |
| 4417 | USA | United States of America | 3523 | Nevada | POLYGON ((-114.04020 37.00313, -114.04025 36.9... |
| 4418 | USA | United States of America | 3524 | New Mexico | POLYGON ((-103.00024 36.50020, -103.04211 36.4... |
| 4419 | USA | United States of America | 3525 | Oregon | MULTIPOLYGON (((-123.99479 46.22024, -123.9848... |
| 4420 | USA | United States of America | 3526 | Utah | POLYGON ((-113.48151 42.00000, -113.29450 42.0... |
| 4421 | USA | United States of America | 3527 | Wyoming | POLYGON ((-104.05312 43.00037, -104.05300 42.8... |
| 4422 | USA | United States of America | 3528 | Arkansas | POLYGON ((-90.37331 35.98619, -90.30080 35.987... |
| 4423 | USA | United States of America | 3529 | Iowa | POLYGON ((-90.64128 42.51177, -90.65422 42.489... |
| 4424 | USA | United States of America | 3530 | Kansas | POLYGON ((-95.09118 39.86072, -95.06782 39.865... |
| 4425 | USA | United States of America | 3531 | Missouri | POLYGON ((-91.60724 40.50218, -91.59636 40.502... |
| 4426 | USA | United States of America | 3532 | Nebraska | POLYGON ((-102.49553 43.00032, -102.32514 43.0... |
| 4427 | USA | United States of America | 3533 | Oklahoma | POLYGON ((-94.61833 36.49983, -94.59592 36.360... |
| 4428 | USA | United States of America | 3534 | South Dakota | POLYGON ((-103.77807 45.94200, -103.54509 45.9... |
| 4429 | USA | United States of America | 3535 | Louisiana | MULTIPOLYGON (((-90.82771 29.04194, -90.82929 ... |
| 4430 | USA | United States of America | 3536 | Texas | MULTIPOLYGON (((-97.16340 26.11359, -97.16638 ... |
| 4431 | USA | United States of America | 3537 | Connecticut | POLYGON ((-72.80641 42.00699, -72.76330 42.010... |
| 4432 | USA | United States of America | 3538 | New Hampshire | POLYGON ((-70.74409 43.08381, -70.75451 43.058... |
| 4433 | USA | United States of America | 3539 | Rhode Island | MULTIPOLYGON (((-71.57157 41.14977, -71.59955 ... |
| 4434 | USA | United States of America | 3540 | Vermont | POLYGON ((-71.57111 44.57469, -71.58347 44.574... |
| 4435 | USA | United States of America | 3541 | Alabama | MULTIPOLYGON (((-88.09097 30.24470, -88.31127 ... |
| 4436 | USA | United States of America | 3542 | Florida | MULTIPOLYGON (((-82.14965 24.55162, -82.14953 ... |
| 4437 | USA | United States of America | 3543 | Georgia | MULTIPOLYGON (((-81.45421 30.73485, -81.46166 ... |
| 4438 | USA | United States of America | 3544 | Mississippi | MULTIPOLYGON (((-88.94274 30.21381, -88.96931 ... |
| 4439 | USA | United States of America | 3545 | South Carolina | MULTIPOLYGON (((-79.36095 33.00568, -79.36840 ... |
| 4440 | USA | United States of America | 3546 | Illinois | POLYGON ((-87.80104 42.49178, -87.61054 42.491... |
| 4441 | USA | United States of America | 3547 | Indiana | POLYGON ((-87.22778 41.75970, -87.22121 41.759... |
| 4442 | USA | United States of America | 3548 | Kentucky | MULTIPOLYGON (((-89.49438 36.50142, -89.55988 ... |
| 4443 | USA | United States of America | 3549 | North Carolina | MULTIPOLYGON (((-77.94205 33.91682, -77.96239 ... |
| 4444 | USA | United States of America | 3550 | Ohio | POLYGON ((-80.52023 40.64678, -80.53571 40.643... |
| 4445 | USA | United States of America | 3551 | Tennessee | POLYGON ((-87.83911 36.64292, -87.83327 36.644... |
| 4446 | USA | United States of America | 3552 | Virginia | MULTIPOLYGON (((-75.90897 36.54942, -75.90896 ... |
| 4447 | USA | United States of America | 3553 | Wisconsin | POLYGON ((-89.95764 47.28611, -89.98547 47.243... |
| 4448 | USA | United States of America | 3554 | West Virginia | POLYGON ((-80.51940 39.72149, -80.38913 39.721... |
| 4449 | USA | United States of America | 3555 | Delaware | POLYGON ((-75.42446 39.80661, -75.42764 39.803... |
| 4450 | USA | United States of America | 3556 | District of Columbia | POLYGON ((-77.02293 38.80277, -77.02739 38.819... |
| 4451 | USA | United States of America | 3557 | Maryland | MULTIPOLYGON (((-75.98129 37.99284, -75.98977 ... |
| 4452 | USA | United States of America | 3558 | New Jersey | MULTIPOLYGON (((-74.35924 39.40156, -74.39338 ... |
| 4453 | USA | United States of America | 3559 | New York | MULTIPOLYGON (((-73.38952 40.61677, -73.57412 ... |
| 4454 | USA | United States of America | 3560 | Pennsylvania | POLYGON ((-79.76291 41.99986, -79.72656 41.999... |
| 4455 | USA | United States of America | 3561 | Maine | MULTIPOLYGON (((-70.61244 42.98402, -70.60839 ... |
| 4456 | USA | United States of America | 3562 | Michigan | POLYGON ((-84.51230 46.45164, -84.49218 46.456... |
| 4457 | USA | United States of America | 3563 | Alaska | MULTIPOLYGON (((-179.08149 51.28644, -179.0694... |
# remove all non US states
us_geojson = us_geojson[~us_geojson['name'].isin(["Rhode Island", "Alaska", "District of Columbia", "Hawaii"])]
plt.rcParams['figure.figsize'] = [20, 20]
us_geojson.boundary.plot()
<AxesSubplot:>
Remember the geojson information on each state from before? We now turn them all into a FeatureCollection for earth engine.
json_fc = json.loads(us_geojson.loc[:, ["name", "geometry"]].to_json())
json_fc['columns'] = {"key": "String"}
roi_fc = ee.FeatureCollection(json_fc)
Ah yes. Code. You can still detect the residual javascript.
# thanks documentation https://developers.google.com/earth-engine/tutorials/community/extract-raster-values-for-points#zonalstatsfc_params_%E2%87%92_eefeaturecollection
def zonalStats(ic, fc, params):
# Initialize internal params dictionary.
_params = {
"reducer": ee.Reducer.mean(),
"scale": None,
"crs": None,
"bands": None,
"bandsRename": None,
"imgProps": None,
"imgPropsRename": None,
"datetimeName": 'datetime',
"datetimeFormat": 'YYYY-MM-dd HH:mm:ss'
}
# Replace initialized params with provided params.
if (params) :
for param in params :
_params[param] = params[param] or _params[param]
# Set default parameters based on an image representative.
imgRep = ic.first();
nonSystemImgProps = ee.Feature(None).copyProperties(imgRep).propertyNames()
if (not _params['bands']): _params['bands'] = imgRep.bandNames()
if (not _params['bandsRename']): _params['bandsRename'] = _params['bands']
if (not _params['imgProps']): _params['imgProps'] = nonSystemImgProps;
if (not _params['imgPropsRename']): _params['imgPropsRename'] = _params['imgProps']
def reduceRegions_over_IC(img):
# Select bands (optionally rename), set a datetime & timestamp property.
img = ee.Image(img.select(_params['bands'], _params['bandsRename']))\
.set(_params['datetimeName'], img.date().format(_params['datetimeFormat']))\
.set('timestamp', img.get('system:time_start'));
# Define final image property dictionary to set in output features.
propsFrom = ee.List(_params['imgProps']).cat(ee.List([_params['datetimeName'], 'timestamp']));
propsTo = ee.List(_params['imgPropsRename']).cat(ee.List([_params['datetimeName'], 'timestamp']));
imgProps = img.toDictionary(propsFrom).rename(propsFrom, propsTo);
# Subset points that intersect the given image.
fcSub = fc.filterBounds(img.geometry());
# Reduce the image by regions and add metadata to each feature.
# It's very funny how reducer.setOutputs() works. Very cringe, but that's just how to fix ee's reducer autonaming single bands to "mean".
return img.reduceRegions(collection=fc, reducer=_params['reducer'].setOutputs([_params['bands'][0]]),
scale=_params['scale'], crs=_params['crs']).map(lambda f: f.set(imgProps))
# Map the reduceRegions function over the image collection.
results = ic.map(reduceRegions_over_IC).flatten().filter(ee.Filter.notNull(_params['bandsRename']));
return results;
Initialise parameters for the Earth Engine API call.
# define dataset names
datasets = dict();
datasets['CH4'] = {"id":"COPERNICUS/S5P/OFFL/L3_CH4", "bands":["CH4_column_volume_mixing_ratio_dry_air"]}
datasets['SO2'] = {"id":"COPERNICUS/S5P/NRTI/L3_SO2", "bands":["SO2_column_number_density", "SO2_slant_column_number_density"]}
datasets['O3'] = {"id":"COPERNICUS/S5P/NRTI/L3_O3", "bands":["O3_column_number_density", "O3_slant_column_number_density", "O3_effective_temperature"]}
datasets['NO2'] = {"id":"COPERNICUS/S5P/NRTI/L3_NO2", "bands":["NO2_column_number_density", "tropospheric_NO2_column_number_density", "stratospheric_NO2_column_number_density"]}
datasets['HCHO'] = {"id":"COPERNICUS/S5P/NRTI/L3_HCHO", "bands":["tropospheric_HCHO_column_number_density", "tropospheric_HCHO_column_number_density_amf", "HCHO_slant_column_number_density"]}
datasets['CO'] = {"id":"COPERNICUS/S5P/NRTI/L3_CO", "bands":["CO_column_number_density", "H2O_column_number_density"]}
datasets['AER_AI'] = {"id":"COPERNICUS/S5P/NRTI/L3_AER_AI", "bands":["absorbing_aerosol_index"]}
start_date = '2020-01-01'
end_date = '2022-01-01'
I would not recommend running this cell. It took 2 hours. Per file.
for key, value in datasets.items():
ic = ee.ImageCollection(value["id"]).filterDate(start_date, end_date)
results = zonalStats(ic, roi_fc, params={"bands": value["bands"],
"scale":50000,
"bandsRename": value["bands"],
"crs": "EPSG:4326"})
desc = key + ";" + start_date + ";" + end_date
task = ee.batch.Export.table.toDrive(collection=results, description=desc, fileFormat="csv", folder="data")
task.start()
print(key + " task started.")
CH4 task started. SO2 task started. O3 task started. NO2 task started. HCHO task started. CO task started. AER_AI task started.
Fairly straightforward data loading. The files are very large so using vaex is much faster, as they do lazy computation of variables, not storing them in memory like pandas.
def process_exported(file_path, bands):
retrieve_bands = bands.copy()
retrieve_bands.insert(0, "datetime")
retrieve_bands.insert(0, "name")
df = vaex.from_csv(file_path, parse_dates=["datetime"])
return df[retrieve_bands]
Note that all data files were stored in the data/ directory. We now iterate through all the dictionary keys and add the filepaths as a value, based on regex filtering using glob.
root_dir = r"data"
for key in datasets.keys():
regex = "*" + key + "*.csv"
pathname = path.join(root_dir, regex)
datasets[key]["filepath"] = glob(pathname)[0]
print(datasets)
{'CH4': {'id': 'COPERNICUS/S5P/OFFL/L3_CH4', 'bands': ['CH4_column_volume_mixing_ratio_dry_air'], 'filepath': 'data\\CH4;2020-01-01;2022-01-01.csv'}, 'SO2': {'id': 'COPERNICUS/S5P/NRTI/L3_SO2', 'bands': ['SO2_column_number_density', 'SO2_slant_column_number_density'], 'filepath': 'data\\SO2;2020-01-01;2022-01-01.csv'}, 'O3': {'id': 'COPERNICUS/S5P/NRTI/L3_O3', 'bands': ['O3_column_number_density', 'O3_slant_column_number_density', 'O3_effective_temperature'], 'filepath': 'data\\O3;2020-01-01;2022-01-01.csv'}, 'NO2': {'id': 'COPERNICUS/S5P/NRTI/L3_NO2', 'bands': ['NO2_column_number_density', 'tropospheric_NO2_column_number_density', 'stratospheric_NO2_column_number_density'], 'filepath': 'data\\NO2;2020-01-01;2022-01-01.csv'}, 'HCHO': {'id': 'COPERNICUS/S5P/NRTI/L3_HCHO', 'bands': ['tropospheric_HCHO_column_number_density', 'tropospheric_HCHO_column_number_density_amf', 'HCHO_slant_column_number_density'], 'filepath': 'data\\HCHO;2020-01-01;2022-01-01.csv'}, 'CO': {'id': 'COPERNICUS/S5P/NRTI/L3_CO', 'bands': ['CO_column_number_density', 'H2O_column_number_density'], 'filepath': 'data\\CO;2020-01-01;2022-01-01.csv'}, 'AER_AI': {'id': 'COPERNICUS/S5P/NRTI/L3_AER_AI', 'bands': ['absorbing_aerosol_index'], 'filepath': 'data\\AER_AI;2020-01-01;2022-01-01.csv'}}
We then initalise the starting df using AER_AI, and then iterate through all the keys, processing their vaex dataframes using process_exported and merging them to the starting df as we go.
# create the initial df from AER_AI
df = process_exported(datasets['AER_AI']['filepath'], datasets['AER_AI']['bands'])
df = df.groupby([vaex.BinnerTime.per_day(df.datetime), df['name']]).agg({'absorbing_aerosol_index': 'mean'})
df = df.to_pandas_df()
for key in datasets:
if key == 'AER_AI': continue
f = datasets[key]
print(f['id'])
df_tomerge = process_exported(f['filepath'], f["bands"])
# we bin the dates, as most values have multiple readings per state in the same date, we take the average of all the values in the same day
# this reduces the number of values, and prevents duplicates during merging.
agg_dict = dict([band, "mean"] for band in f["bands"])
df_tomerge = df_tomerge.groupby([vaex.BinnerTime.per_day(df_tomerge.datetime), df_tomerge['name']]).agg(agg_dict)
df = df.merge(df_tomerge.to_pandas_df())
df
COPERNICUS/S5P/OFFL/L3_CH4 COPERNICUS/S5P/NRTI/L3_SO2 COPERNICUS/S5P/NRTI/L3_O3 COPERNICUS/S5P/NRTI/L3_NO2 COPERNICUS/S5P/NRTI/L3_HCHO COPERNICUS/S5P/NRTI/L3_CO
| datetime | name | absorbing_aerosol_index | CH4_column_volume_mixing_ratio_dry_air | SO2_column_number_density | SO2_slant_column_number_density | O3_column_number_density | O3_slant_column_number_density | O3_effective_temperature | NO2_column_number_density | tropospheric_NO2_column_number_density | stratospheric_NO2_column_number_density | tropospheric_HCHO_column_number_density | tropospheric_HCHO_column_number_density_amf | HCHO_slant_column_number_density | CO_column_number_density | H2O_column_number_density | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2020-01-04 | California | -1.302309 | 1877.423259 | 0.000839 | 0.000143 | 0.131675 | 0.434540 | 228.558268 | 0.000065 | 0.000036 | 0.000029 | 0.000029 | 0.978985 | -0.000008 | 0.029724 | 476.676151 |
| 1 | 2020-01-06 | Tennessee | -1.364598 | 1847.756329 | 0.000507 | 0.000167 | 0.130055 | 0.399064 | 225.298974 | 0.000069 | 0.000045 | 0.000024 | 0.000064 | 1.236318 | 0.000040 | 0.032176 | 473.255912 |
| 2 | 2020-01-07 | Mississippi | -1.569424 | 1857.628257 | 0.000527 | 0.000140 | 0.124243 | 0.379138 | 226.335102 | 0.000058 | 0.000030 | 0.000027 | 0.000069 | 0.917662 | 0.000014 | 0.032303 | 352.338888 |
| 3 | 2020-01-22 | New Jersey | -1.312634 | 1847.811467 | 0.001519 | 0.000244 | 0.153461 | 0.601108 | 222.785718 | 0.000135 | 0.000109 | 0.000026 | 0.000030 | 1.213370 | 0.000010 | 0.040528 | 368.570551 |
| 4 | 2020-01-25 | Alabama | -1.134544 | 1831.286644 | 0.000993 | 0.000197 | 0.135140 | 0.508267 | 222.340566 | 0.000059 | 0.000036 | 0.000028 | 0.000062 | 1.181874 | 0.000027 | 0.032801 | 639.293485 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 19101 | 2021-12-16 | Mississippi | -1.197653 | 1812.727961 | 0.000092 | 0.000054 | 0.116146 | 0.357491 | 227.363749 | 0.000052 | 0.000022 | 0.000028 | 0.000031 | 1.504923 | -0.000008 | 0.025573 | 1618.674073 |
| 19102 | 2021-12-24 | Florida | 0.414362 | 1887.601321 | 0.000956 | 0.000169 | 0.118702 | 0.453879 | 224.369659 | 0.000060 | 0.000024 | 0.000036 | 0.000083 | 1.108676 | 0.000042 | 0.029374 | 906.245482 |
| 19103 | 2021-12-24 | Arizona | -0.075089 | 1924.671767 | 0.001928 | 0.000427 | 0.129865 | 0.520854 | 222.640916 | 0.000043 | 0.000012 | 0.000030 | 0.000046 | 1.172407 | 0.000008 | 0.022212 | 723.149736 |
| 19104 | 2021-12-25 | Georgia | 0.395280 | 1865.095538 | 0.000478 | 0.000126 | 0.114920 | 0.440511 | 224.026752 | 0.000053 | 0.000014 | 0.000034 | 0.000059 | 1.080180 | 0.000016 | 0.028374 | 1224.921932 |
| 19105 | 2021-12-31 | Arizona | -0.721039 | 1883.382942 | 0.000361 | 0.000148 | 0.133554 | 0.418152 | 224.518795 | 0.000044 | 0.000013 | 0.000032 | -0.000013 | 1.352835 | -0.000055 | 0.023580 | 608.668468 |
19106 rows × 17 columns
df.info()
<class 'pandas.core.frame.DataFrame'> Int64Index: 19106 entries, 0 to 19105 Data columns (total 17 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 datetime 19106 non-null datetime64[ns] 1 name 19106 non-null object 2 absorbing_aerosol_index 19106 non-null float64 3 CH4_column_volume_mixing_ratio_dry_air 19106 non-null float64 4 SO2_column_number_density 19106 non-null float64 5 SO2_slant_column_number_density 19106 non-null float64 6 O3_column_number_density 19106 non-null float64 7 O3_slant_column_number_density 19106 non-null float64 8 O3_effective_temperature 19106 non-null float64 9 NO2_column_number_density 19106 non-null float64 10 tropospheric_NO2_column_number_density 19106 non-null float64 11 stratospheric_NO2_column_number_density 19106 non-null float64 12 tropospheric_HCHO_column_number_density 19106 non-null float64 13 tropospheric_HCHO_column_number_density_amf 19106 non-null float64 14 HCHO_slant_column_number_density 19106 non-null float64 15 CO_column_number_density 19106 non-null float64 16 H2O_column_number_density 19106 non-null float64 dtypes: datetime64[ns](1), float64(15), object(1) memory usage: 2.6+ MB
We now set the datetime and name columns to be our indices, and sort our data to follow based on datetime followed by country. We then rename the columns to shorter, more easy-to-manipulate names, based on the following one-to-one table:
| Before | After |
|---|---|
absorbing_aerosol_index |
UVAI |
CH4_column_volume_mixing_ratio_dry_air |
CH4 |
CO_column_number_density |
CO |
H2O_column_number_density |
H2O |
tropospheric_HCHO_column_number_density |
tropoHCHO |
tropospheric_HCHO_column_number_density_amf |
tropoHCHOamf |
HCHO_slant_column_number_density |
slantHCHO |
NO2_column_number_density |
NO2 |
tropospheric_NO2_column_number_density |
tropoNO2 |
stratospheric_NO2_column_number_density |
stratoNO2 |
O3_column_number_density |
O3 |
O3_slant_column_number_density |
slantO3 |
O3_effective_temperature |
O3Teff |
SO2_column_number_density |
SO2 |
SO2_slant_column_number_density |
slantSO2 |
df = df.set_index(["datetime", "name"]).sort_index().rename(columns={"absorbing_aerosol_index":"UVAI", "CH4_column_volume_mixing_ratio_dry_air":"CH4", "CO_column_number_density":"CO", "H2O_column_number_density":"H2O", "tropospheric_HCHO_column_number_density":"tropoHCHO", "tropospheric_HCHO_column_number_density_amf":"trompoHCHOamf", "HCHO_slant_column_number_density":"slantHCHO", "NO2_column_number_density":"NO2", "tropospheric_NO2_column_number_density":"tropoNO2", "stratospheric_NO2_column_number_density":"stratoNO2", "O3_column_number_density":"O3", "O3_slant_column_number_density":"slantO3", "O3_effective_temperature":"O3Teff", "SO2_column_number_density":"SO2", "SO2_slant_column_number_density":"slantSO2"})
df
| UVAI | CH4 | SO2 | slantSO2 | O3 | slantO3 | O3Teff | NO2 | tropoNO2 | stratoNO2 | tropoHCHO | trompoHCHOamf | slantHCHO | CO | H2O | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| datetime | name | |||||||||||||||
| 2020-01-01 | Alabama | -1.857279 | 1851.422663 | 0.000634 | 0.000162 | 0.131059 | 0.376028 | 224.267768 | 0.000048 | 0.000019 | 0.000026 | 0.000064 | 1.002517 | 0.000020 | 0.032068 | 617.789468 |
| Arizona | -1.501533 | 1862.697229 | 0.001869 | 0.000331 | 0.141398 | 0.428382 | 226.678055 | 0.000052 | 0.000022 | 0.000026 | 0.000041 | 1.061395 | 0.000004 | 0.026534 | 684.330827 | |
| Arkansas | -1.641902 | 1873.183618 | 0.000667 | 0.000166 | 0.145351 | 0.459384 | 224.733548 | 0.000051 | 0.000020 | 0.000026 | 0.000087 | 0.851757 | 0.000029 | 0.030415 | 524.404399 | |
| California | -0.911412 | 1883.150108 | 0.001011 | 0.000190 | 0.137392 | 0.512113 | 228.147282 | 0.000065 | 0.000044 | 0.000025 | 0.000067 | 1.091332 | 0.000030 | 0.029348 | 897.880270 | |
| Florida | -1.617711 | 1840.478564 | 0.000197 | 0.000058 | 0.115436 | 0.325569 | 224.439818 | 0.000060 | 0.000033 | 0.000027 | 0.000070 | 0.998220 | 0.000022 | 0.031240 | 1389.138246 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-12-31 | Louisiana | -1.231453 | 1877.135982 | 0.000118 | 0.000077 | 0.124393 | 0.358781 | 228.041266 | 0.000044 | 0.000014 | 0.000032 | 0.000081 | 1.376686 | 0.000061 | 0.028789 | 2001.649260 |
| New Mexico | -0.167673 | 1876.840170 | 0.000286 | 0.000076 | 0.129040 | 0.474315 | 221.275302 | 0.000042 | 0.000008 | 0.000032 | -0.000013 | 1.157179 | -0.000049 | 0.022373 | 704.473791 | |
| North Carolina | 0.438441 | 1855.335995 | 0.002123 | 0.000359 | 0.140183 | 0.568000 | 224.734080 | 0.000045 | 0.000012 | 0.000035 | 0.000075 | 1.017024 | 0.000036 | 0.029267 | 1275.679924 | |
| South Carolina | 0.198841 | 1858.691068 | 0.002760 | 0.000403 | 0.136064 | 0.525551 | 226.101428 | 0.000046 | 0.000011 | 0.000034 | 0.000063 | 0.971292 | 0.000018 | 0.028811 | 1646.595774 | |
| Texas | -0.070929 | 1879.023884 | 0.000083 | 0.000026 | 0.131176 | 0.499624 | 222.008966 | 0.000047 | 0.000026 | 0.000033 | 0.000133 | 0.809671 | 0.000058 | 0.024562 | 1467.127803 |
19106 rows × 15 columns
Normalise values to between 0 and 1.
dfrange = df.max()-df.min()
dfmin = df.min()
values = pd.DataFrame({"range":dfrange, "min":dfmin})
values
| range | min | |
|---|---|---|
| UVAI | 8.655660 | -2.977895 |
| CH4 | 251.481256 | 1716.521507 |
| SO2 | 0.013002 | -0.000995 |
| slantSO2 | 0.003373 | -0.000836 |
| O3 | 0.114978 | 0.100792 |
| slantO3 | 0.551211 | 0.255271 |
| O3Teff | 31.443662 | 207.011662 |
| NO2 | 0.000171 | 0.000013 |
| tropoNO2 | 0.000195 | -0.000019 |
| stratoNO2 | 0.000054 | 0.000019 |
| tropoHCHO | 0.001127 | -0.000387 |
| trompoHCHOamf | 2.270414 | 0.547066 |
| slantHCHO | 0.001029 | -0.000410 |
| CO | 0.119473 | 0.018954 |
| H2O | 9798.182682 | 71.447735 |
df = (df-df.min())/(df.max()-df.min())
df
| UVAI | CH4 | SO2 | slantSO2 | O3 | slantO3 | O3Teff | NO2 | tropoNO2 | stratoNO2 | tropoHCHO | trompoHCHOamf | slantHCHO | CO | H2O | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| datetime | name | |||||||||||||||
| 2020-01-01 | Alabama | 0.129466 | 0.536426 | 0.125310 | 0.295915 | 0.263242 | 0.219076 | 0.548794 | 0.202752 | 0.196354 | 0.120865 | 0.399629 | 0.200602 | 0.418022 | 0.109763 | 0.055759 |
| Arizona | 0.170566 | 0.581259 | 0.220250 | 0.346169 | 0.353167 | 0.314057 | 0.625449 | 0.225444 | 0.210016 | 0.115512 | 0.379465 | 0.226535 | 0.402161 | 0.063444 | 0.062551 | |
| Arkansas | 0.154349 | 0.622957 | 0.127808 | 0.297120 | 0.387543 | 0.370300 | 0.563608 | 0.223675 | 0.201385 | 0.118010 | 0.419969 | 0.134201 | 0.426246 | 0.095927 | 0.046229 | |
| California | 0.238744 | 0.662589 | 0.154266 | 0.304132 | 0.318321 | 0.465959 | 0.672174 | 0.301876 | 0.324919 | 0.110159 | 0.402795 | 0.239721 | 0.427957 | 0.086998 | 0.084345 | |
| Florida | 0.157144 | 0.492908 | 0.091659 | 0.265213 | 0.127362 | 0.127534 | 0.554266 | 0.271363 | 0.268341 | 0.134711 | 0.405254 | 0.198710 | 0.419529 | 0.102836 | 0.134483 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-12-31 | Louisiana | 0.201769 | 0.638674 | 0.085562 | 0.270712 | 0.205266 | 0.187787 | 0.668803 | 0.179660 | 0.169056 | 0.232619 | 0.414583 | 0.365404 | 0.457577 | 0.082317 | 0.196996 |
| New Mexico | 0.324669 | 0.637497 | 0.098525 | 0.270342 | 0.245684 | 0.397388 | 0.453625 | 0.166294 | 0.141015 | 0.233960 | 0.331613 | 0.268723 | 0.350829 | 0.028620 | 0.064606 | |
| North Carolina | 0.394694 | 0.551987 | 0.239815 | 0.354422 | 0.342600 | 0.567349 | 0.563625 | 0.182806 | 0.158877 | 0.289233 | 0.409270 | 0.206992 | 0.433742 | 0.086320 | 0.122904 | |
| South Carolina | 0.367013 | 0.565329 | 0.288756 | 0.367386 | 0.306770 | 0.490338 | 0.607110 | 0.191033 | 0.157030 | 0.260059 | 0.398835 | 0.186849 | 0.415874 | 0.082503 | 0.160759 | |
| Texas | 0.335846 | 0.646181 | 0.082884 | 0.255527 | 0.264259 | 0.443302 | 0.476958 | 0.199479 | 0.233341 | 0.245947 | 0.461080 | 0.115664 | 0.454958 | 0.046942 | 0.142443 |
19106 rows × 15 columns
We now perform Correlation calculations to analyse if any variable can be simply eliminated, or in what ways they are correlated. We do this by computing the correlation matrix, as stored in a variable corrmat. Following this, we plot a Heatmap of the Correlation Matrix, as shown below.
corrmat = df.corr()
corrmat
| UVAI | CH4 | SO2 | slantSO2 | O3 | slantO3 | O3Teff | NO2 | tropoNO2 | stratoNO2 | tropoHCHO | trompoHCHOamf | slantHCHO | CO | H2O | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UVAI | 1.000000 | 0.279078 | 0.054034 | 0.041041 | -0.248161 | 0.103640 | -0.077454 | -0.068385 | -0.042478 | -0.039923 | 0.016303 | 0.128532 | 0.036201 | 0.146688 | -0.128833 |
| CH4 | 0.279078 | 1.000000 | 0.012574 | 0.026239 | -0.326540 | -0.072790 | -0.188824 | -0.040042 | 0.094744 | -0.152457 | 0.009228 | -0.052045 | -0.009125 | -0.074105 | -0.169346 |
| SO2 | 0.054034 | 0.012574 | 1.000000 | 0.879411 | 0.144258 | 0.546959 | -0.461599 | -0.135823 | 0.205587 | -0.460530 | -0.282791 | -0.062560 | -0.248754 | 0.015661 | -0.335744 |
| slantSO2 | 0.041041 | 0.026239 | 0.879411 | 1.000000 | 0.172700 | 0.580186 | -0.466886 | -0.197638 | 0.132104 | -0.459589 | -0.324204 | -0.054104 | -0.291720 | -0.018488 | -0.367271 |
| O3 | -0.248161 | -0.326540 | 0.144258 | 0.172700 | 1.000000 | 0.528470 | 0.044809 | 0.009391 | -0.125941 | 0.182268 | -0.316704 | 0.104519 | -0.250725 | 0.210366 | -0.261063 |
| slantO3 | 0.103640 | -0.072790 | 0.546959 | 0.580186 | 0.528470 | 1.000000 | -0.453947 | -0.243817 | 0.006019 | -0.377022 | -0.459103 | 0.146192 | -0.378417 | 0.045733 | -0.475795 |
| O3Teff | -0.077454 | -0.188824 | -0.461599 | -0.466886 | 0.044809 | -0.453947 | 1.000000 | 0.326677 | -0.223764 | 0.802217 | 0.397343 | 0.048578 | 0.319301 | 0.131174 | 0.511871 |
| NO2 | -0.068385 | -0.040042 | -0.135823 | -0.197638 | 0.009391 | -0.243817 | 0.326677 | 1.000000 | 0.706027 | 0.402623 | 0.344234 | -0.089539 | 0.322351 | 0.289165 | 0.373718 |
| tropoNO2 | -0.042478 | 0.094744 | 0.205587 | 0.132104 | -0.125941 | 0.006019 | -0.223764 | 0.706027 | 1.000000 | -0.271077 | 0.124799 | -0.075248 | 0.148961 | 0.199298 | 0.099374 |
| stratoNO2 | -0.039923 | -0.152457 | -0.460530 | -0.459589 | 0.182268 | -0.377022 | 0.802217 | 0.402623 | -0.271077 | 1.000000 | 0.350471 | -0.053850 | 0.281873 | 0.127190 | 0.384429 |
| tropoHCHO | 0.016303 | 0.009228 | -0.282791 | -0.324204 | -0.316704 | -0.459103 | 0.397343 | 0.344234 | 0.124799 | 0.350471 | 1.000000 | -0.180120 | 0.938336 | 0.107442 | 0.571252 |
| trompoHCHOamf | 0.128532 | -0.052045 | -0.062560 | -0.054104 | 0.104519 | 0.146192 | 0.048578 | -0.089539 | -0.075248 | -0.053850 | -0.180120 | 1.000000 | 0.038274 | -0.049385 | 0.077954 |
| slantHCHO | 0.036201 | -0.009125 | -0.248754 | -0.291720 | -0.250725 | -0.378417 | 0.319301 | 0.322351 | 0.148961 | 0.281873 | 0.938336 | 0.038274 | 1.000000 | 0.135181 | 0.519513 |
| CO | 0.146688 | -0.074105 | 0.015661 | -0.018488 | 0.210366 | 0.045733 | 0.131174 | 0.289165 | 0.199298 | 0.127190 | 0.107442 | -0.049385 | 0.135181 | 1.000000 | 0.026833 |
| H2O | -0.128833 | -0.169346 | -0.335744 | -0.367271 | -0.261063 | -0.475795 | 0.511871 | 0.373718 | 0.099374 | 0.384429 | 0.571252 | 0.077954 | 0.519513 | 0.026833 | 1.000000 |
plt.figure(figsize=(15, 12))
sns.heatmap(corrmat, cmap="RdBu_r", vmin=-1, vmax=1, annot=True)
<AxesSubplot:>
We note that the correlation between the Effective Temperature at Ozone and the Stratospheric NO2 Column Density are incredibly low, hence we plot between the two datasets to search for some form of correlation, as shown below:
plt.figure(figsize=(16,16))
sns.regplot(df.O3Teff, df.stratoNO2, scatter_kws=dict(alpha=0.2), line_kws=dict(linestyle="--"))
plt.gca().set(title="Plot of Stratospheric $NO_2$ Column Density against Effective Temperature of $O_3$", xlabel="Effective Temperature of $O_3$", ylabel="Stratospheric $NO_2$ Column Density")
C:\Users\Prannaya\.conda\envs\analytics\lib\site-packages\seaborn\_decorators.py:36: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation. warnings.warn(
[Text(0.5, 1.0, 'Plot of Stratospheric $NO_2$ Column Density against Effective Temperature of $O_3$'), Text(0.5, 0, 'Effective Temperature of $O_3$'), Text(0, 0.5, 'Stratospheric $NO_2$ Column Density')]
df = df.reset_index(level=1)
df
| name | UVAI | CH4 | SO2 | slantSO2 | O3 | slantO3 | O3Teff | NO2 | tropoNO2 | stratoNO2 | tropoHCHO | trompoHCHOamf | slantHCHO | CO | H2O | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| datetime | ||||||||||||||||
| 2020-01-01 | Alabama | 0.129466 | 0.536426 | 0.125310 | 0.295915 | 0.263242 | 0.219076 | 0.548794 | 0.202752 | 0.196354 | 0.120865 | 0.399629 | 0.200602 | 0.418022 | 0.109763 | 0.055759 |
| 2020-01-01 | Arizona | 0.170566 | 0.581259 | 0.220250 | 0.346169 | 0.353167 | 0.314057 | 0.625449 | 0.225444 | 0.210016 | 0.115512 | 0.379465 | 0.226535 | 0.402161 | 0.063444 | 0.062551 |
| 2020-01-01 | Arkansas | 0.154349 | 0.622957 | 0.127808 | 0.297120 | 0.387543 | 0.370300 | 0.563608 | 0.223675 | 0.201385 | 0.118010 | 0.419969 | 0.134201 | 0.426246 | 0.095927 | 0.046229 |
| 2020-01-01 | California | 0.238744 | 0.662589 | 0.154266 | 0.304132 | 0.318321 | 0.465959 | 0.672174 | 0.301876 | 0.324919 | 0.110159 | 0.402795 | 0.239721 | 0.427957 | 0.086998 | 0.084345 |
| 2020-01-01 | Florida | 0.157144 | 0.492908 | 0.091659 | 0.265213 | 0.127362 | 0.127534 | 0.554266 | 0.271363 | 0.268341 | 0.134711 | 0.405254 | 0.198710 | 0.419529 | 0.102836 | 0.134483 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-12-31 | Louisiana | 0.201769 | 0.638674 | 0.085562 | 0.270712 | 0.205266 | 0.187787 | 0.668803 | 0.179660 | 0.169056 | 0.232619 | 0.414583 | 0.365404 | 0.457577 | 0.082317 | 0.196996 |
| 2021-12-31 | New Mexico | 0.324669 | 0.637497 | 0.098525 | 0.270342 | 0.245684 | 0.397388 | 0.453625 | 0.166294 | 0.141015 | 0.233960 | 0.331613 | 0.268723 | 0.350829 | 0.028620 | 0.064606 |
| 2021-12-31 | North Carolina | 0.394694 | 0.551987 | 0.239815 | 0.354422 | 0.342600 | 0.567349 | 0.563625 | 0.182806 | 0.158877 | 0.289233 | 0.409270 | 0.206992 | 0.433742 | 0.086320 | 0.122904 |
| 2021-12-31 | South Carolina | 0.367013 | 0.565329 | 0.288756 | 0.367386 | 0.306770 | 0.490338 | 0.607110 | 0.191033 | 0.157030 | 0.260059 | 0.398835 | 0.186849 | 0.415874 | 0.082503 | 0.160759 |
| 2021-12-31 | Texas | 0.335846 | 0.646181 | 0.082884 | 0.255527 | 0.264259 | 0.443302 | 0.476958 | 0.199479 | 0.233341 | 0.245947 | 0.461080 | 0.115664 | 0.454958 | 0.046942 | 0.142443 |
19106 rows × 16 columns
plt.figure(figsize=(16,32))
sns.stripplot(data=df, y="name", x="UVAI").set(xlabel="(Scaled) UV Absorbing Index", ylabel="Name of State", title="(Scaled) UV Absorbing Index based on the Number of States")
[Text(0.5, 0, '(Scaled) UV Absorbing Index'), Text(0, 0.5, 'Name of State'), Text(0.5, 1.0, '(Scaled) UV Absorbing Index based on the Number of States')]
plt.figure(figsize=(16,32))
sns.boxplot(data=df, y="name", x="UVAI").set(xlabel="(Scaled) UV Absorbing Index", ylabel="Name of State", title="Boxplot of Scaled UV Absorbing Index based on the Number of States")
[Text(0.5, 0, '(Scaled) UV Absorbing Index'), Text(0, 0.5, 'Name of State'), Text(0.5, 1.0, 'Boxplot of Scaled UV Absorbing Index based on the Number of States')]
plt.figure(figsize=(16,32))
sns.violinplot(data=df, y="name", x="UVAI").set(xlabel="(Scaled) UV Absorbing Index", ylabel="Name of State", title="Violin Plot of (Scaled) UV Absorbing Index based on the Number of States")
[Text(0.5, 0, '(Scaled) UV Absorbing Index'), Text(0, 0.5, 'Name of State'), Text(0.5, 1.0, 'Violin Plot of (Scaled) UV Absorbing Index based on the Number of States')]
That is to say, converting the data into Tensors in Numpy.
names = pd.get_dummies(df.name)
names
| Alabama | Arizona | Arkansas | California | Colorado | Connecticut | Delaware | Florida | Georgia | Idaho | ... | South Dakota | Tennessee | Texas | Utah | Vermont | Virginia | Washington | West Virginia | Wisconsin | Wyoming | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| datetime | |||||||||||||||||||||
| 2020-01-01 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-01 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-01 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-01 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-12-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2021-12-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2021-12-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2021-12-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2021-12-31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
19106 rows × 47 columns
df.loc[:, names.columns] = names
df = df.drop(columns="name").reset_index().drop(columns="datetime")
df
| UVAI | CH4 | SO2 | slantSO2 | O3 | slantO3 | O3Teff | NO2 | tropoNO2 | stratoNO2 | ... | South Dakota | Tennessee | Texas | Utah | Vermont | Virginia | Washington | West Virginia | Wisconsin | Wyoming | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.129466 | 0.536426 | 0.125310 | 0.295915 | 0.263242 | 0.219076 | 0.548794 | 0.202752 | 0.196354 | 0.120865 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0.170566 | 0.581259 | 0.220250 | 0.346169 | 0.353167 | 0.314057 | 0.625449 | 0.225444 | 0.210016 | 0.115512 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0.154349 | 0.622957 | 0.127808 | 0.297120 | 0.387543 | 0.370300 | 0.563608 | 0.223675 | 0.201385 | 0.118010 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0.238744 | 0.662589 | 0.154266 | 0.304132 | 0.318321 | 0.465959 | 0.672174 | 0.301876 | 0.324919 | 0.110159 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0.157144 | 0.492908 | 0.091659 | 0.265213 | 0.127362 | 0.127534 | 0.554266 | 0.271363 | 0.268341 | 0.134711 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 19101 | 0.201769 | 0.638674 | 0.085562 | 0.270712 | 0.205266 | 0.187787 | 0.668803 | 0.179660 | 0.169056 | 0.232619 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19102 | 0.324669 | 0.637497 | 0.098525 | 0.270342 | 0.245684 | 0.397388 | 0.453625 | 0.166294 | 0.141015 | 0.233960 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19103 | 0.394694 | 0.551987 | 0.239815 | 0.354422 | 0.342600 | 0.567349 | 0.563625 | 0.182806 | 0.158877 | 0.289233 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19104 | 0.367013 | 0.565329 | 0.288756 | 0.367386 | 0.306770 | 0.490338 | 0.607110 | 0.191033 | 0.157030 | 0.260059 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19105 | 0.335846 | 0.646181 | 0.082884 | 0.255527 | 0.264259 | 0.443302 | 0.476958 | 0.199479 | 0.233341 | 0.245947 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
19106 rows × 62 columns
X = df.loc[:, "CH4":].values
y = df.UVAI.values
X, y
(array([[0.53642629, 0.12530975, 0.29591486, ..., 0. , 0. ,
0. ],
[0.58125892, 0.22025031, 0.3461687 , ..., 0. , 0. ,
0. ],
[0.62295741, 0.127808 , 0.29712036, ..., 0. , 0. ,
0. ],
...,
[0.55198741, 0.23981461, 0.35442181, ..., 0. , 0. ,
0. ],
[0.56532866, 0.28875619, 0.3673858 , ..., 0. , 0. ,
0. ],
[0.64618087, 0.08288379, 0.25552659, ..., 0. , 0. ,
0. ]]),
array([0.12946628, 0.17056608, 0.15434906, ..., 0.39469384, 0.36701251,
0.33584569]))
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 420691337)
def computeValues(model, X, y):
"""
Computes the following values based on a given model, X and y:
- R^2 Value (R2)
- Mean Squared Error (MSE)
- Mean Absolute Error (MAE)
- Mean Squared Log Error (MSLE)
- Median Absolute Error (MdAE)
- Mean Absolute Percentage Error (MAPE)
"""
h = model.predict(X)
values = pd.Series({
"R2":model.score(X, y),
"MSE":mean_squared_error(y, h),
"MAE":mean_absolute_error(y, h),
"MdAE":median_absolute_error(y, h),
"MAPE":mean_absolute_percentage_error(y, h)
})
if np.all(y > 0) and np.all(h > 0): values["MSLE"] = mean_squared_log_error(y, h)
return values
def yhplot(y_test, ys, labels):
fig, axes = plt.subplots(2, figsize=(16,32))
axes[0].plot(y_test, y_test, label="Original $y$", alpha=0.4)
for y_pred, label in zip(ys, labels):
axes[0].scatter(y_test, y_pred, label=label, alpha=0.4)
axes[0].legend()
axes[0].set(xlabel="Expected Value of UVAI", ylabel="Predicted Value of UVAI", title="Plot of Expected against Predicted Values of UVAI")
sns.kdeplot(y_test, label="Original $y$", ax=axes[1])
for y_pred, label in zip(ys, labels):
sns.kdeplot(y_pred, label=label, ax=axes[1])
axes[1].legend()
axes[1].set(xlabel="Scaled UVAI Values", title="KDE Plot of Expected and Predicted Values of UVAI")
Based on the R2 value of 0.3942, there is weak correlation between the input variables and output variables when using an MLRM. Plotting the prediction values, we can see that MLRM does not produce satisfactory results.
mlrm = LinearRegression()
mlrm.fit(X_train, y_train)
y_mlrm = mlrm.predict(X_test)
y_mlrm
array([0.27896118, 0.17984009, 0.24465942, ..., 0.25628662, 0.23840332,
0.12261963])
computeValues(mlrm, X_test, y_test)
R2 0.394192 MSE 0.004907 MAE 0.056199 MdAE 0.048838 MAPE 0.281654 MSLE 0.003085 dtype: float64
yhplot(y_test, [y_mlrm], ["MLRM"])
Based on the R2 value of 0.3926, there is similarly weak relationship between the input and output values for Ridge regression model. Plotting the predicted values, we can see that Ridge regression does not produce satisfactory results.
ridge = Ridge(alpha=1.0)
ridge.fit(X_train, y_train)
y_ridge = ridge.predict(X_test)
y_ridge
array([0.27475558, 0.1806058 , 0.24028131, ..., 0.25444223, 0.23526421,
0.1210801 ])
computeValues(ridge, X_test, y_test)
R2 0.392643 MSE 0.004920 MAE 0.056221 MdAE 0.048979 MAPE 0.281714 MSLE 0.003088 dtype: float64
yhplot(y_test, [y_ridge], ["Ridge"])
Given that the R2 is negative, we can clearly see that the fit of the variables is probably better off explained with a horizontal line than the Lasso regression model.
lasso = Lasso(alpha=1.0)
lasso.fit(X_train, y_train)
y_lasso = lasso.predict(X_test)
y_lasso
array([0.22287857, 0.22287857, 0.22287857, ..., 0.22287857, 0.22287857,
0.22287857])
computeValues(lasso, X_test, y_test)
R2 -0.000435 MSE 0.008104 MAE 0.068700 MdAE 0.056339 MAPE 0.358104 MSLE 0.005044 dtype: float64
yhplot(y_test, [y_lasso], ["Lasso"])
Support Vector Regression normally does not scale well to datasets beyond 10,000 samples (we have nearly 19,106) due to its quadratic nature, however we can afford to wait the time it takes to compute. It seems that RBF and Polynomial kernels perform the best, even compared to previous models.
svr_rbf = SVR(kernel="rbf", C=100, gamma=0.1, epsilon=0.1)
svr_lin = SVR(kernel="linear", C=100, gamma="auto")
svr_poly = SVR(kernel="poly", C=100, gamma="auto", degree=3, epsilon=0.1, coef0=1)
svrs = [svr_rbf, svr_lin, svr_poly]
kernel_label = ["RBF", "Linear", "Polynomial"]
y_svrs = []
for ix, svr in enumerate(svrs):
print(kernel_label[ix])
svr.fit(X_train, y_train)
y_svr = svr.predict(X_test)
y_svrs.append(y_svr)
RBF Linear Polynomial
pd.DataFrame(dict(zip(kernel_label, map(lambda svr: computeValues(svr, X_test, y_test), svrs)))).T
| MAE | MAPE | MSE | MSLE | MdAE | R2 | |
|---|---|---|---|---|---|---|
| RBF | 0.052162 | 0.271719 | 0.004029 | NaN | 0.047338 | 0.502677 |
| Linear | 0.059769 | 0.314378 | 0.005142 | 0.003268 | 0.056179 | 0.365180 |
| Polynomial | 0.055835 | 0.292793 | 0.004509 | NaN | 0.052141 | 0.443391 |
yhplot(y_test, y_svrs, kernel_label)
Absolutely terrible results.
rf = RandomForestRegressor(max_depth=2, random_state=0)
rf.fit(X_train, y_train)
y_rf = rf.predict(X_test)
y_rf
array([0.21441084, 0.20931744, 0.28617961, ..., 0.25632391, 0.21907803,
0.20213858])
computeValues(rf, X_test, y_test)
R2 0.225575 MSE 0.006273 MAE 0.061141 MdAE 0.049370 MAPE 0.320980 MSLE 0.003959 dtype: float64
yhplot(y_test, [y_rf], ["Random Forest"])
results = pd.DataFrame(dict(zip(["MLRM","Ridge","Lasso","RBF Kernel SVR","Linear Kernel SVR","Polynomial Kernel SVR", "Random Forest"], map(lambda clf: computeValues(clf, X_test, y_test), [mlrm, ridge, lasso]+svrs+[rf])))).T
results
| MAE | MAPE | MSE | MSLE | MdAE | R2 | |
|---|---|---|---|---|---|---|
| MLRM | 0.056199 | 0.281654 | 0.004907 | 0.003085 | 0.048838 | 0.394192 |
| Ridge | 0.056221 | 0.281714 | 0.004920 | 0.003088 | 0.048979 | 0.392643 |
| Lasso | 0.068700 | 0.358104 | 0.008104 | 0.005044 | 0.056339 | -0.000435 |
| RBF Kernel SVR | 0.052162 | 0.271719 | 0.004029 | NaN | 0.047338 | 0.502677 |
| Linear Kernel SVR | 0.059769 | 0.314378 | 0.005142 | 0.003268 | 0.056179 | 0.365180 |
| Polynomial Kernel SVR | 0.055835 | 0.292793 | 0.004509 | NaN | 0.052141 | 0.443391 |
| Random Forest | 0.061141 | 0.320980 | 0.006273 | 0.003959 | 0.049370 | 0.225575 |
results.sort_values("R2", ascending=False)
| MAE | MAPE | MSE | MSLE | MdAE | R2 | |
|---|---|---|---|---|---|---|
| RBF Kernel SVR | 0.052162 | 0.271719 | 0.004029 | NaN | 0.047338 | 0.502677 |
| Polynomial Kernel SVR | 0.055835 | 0.292793 | 0.004509 | NaN | 0.052141 | 0.443391 |
| MLRM | 0.056199 | 0.281654 | 0.004907 | 0.003085 | 0.048838 | 0.394192 |
| Ridge | 0.056221 | 0.281714 | 0.004920 | 0.003088 | 0.048979 | 0.392643 |
| Linear Kernel SVR | 0.059769 | 0.314378 | 0.005142 | 0.003268 | 0.056179 | 0.365180 |
| Random Forest | 0.061141 | 0.320980 | 0.006273 | 0.003959 | 0.049370 | 0.225575 |
| Lasso | 0.068700 | 0.358104 | 0.008104 | 0.005044 | 0.056339 | -0.000435 |
Based on these values, we note that the RBF Kernel SVR is the best model. We continue to visualise this data as shown below. Since Lasso is very clearly a bad fit for the datapoints, we exclude the predictions from Lasso Regression from comparison with other models.
ys = [y_mlrm, y_ridge, *y_svrs, y_rf]
yhplot(y_test, ys, ["MLRM","Ridge","RBF Kernel SVR","Linear Kernel SVR","Polynomial Kernel SVR", "Random Forest"])
These plots seem to make it all the more clear that the RBF Kernel SVR is the most accurate model, due to it's higher R2 and lower MSPE values.
In conclusion, the best performing model is the RBF Kernel SVR due to the fact that they have a higher R2 and lowest mean squared error compared to the other models. Note that it seems the R2 values are very low, the data seems to be quite unrelated to the UVAI. Hence, gas column densities and one hot encoded locations may not be an optimal set of predictor variables for predicting UVAI.
Due to the low order data, all models trained relatively quickly, and considering speed as a factor in determining the best model is unnecessary.
For future work, perhaps more different variables could be considered when predicting UVAI.